Mapas coropléticos sobre la sequía en EEUU con R
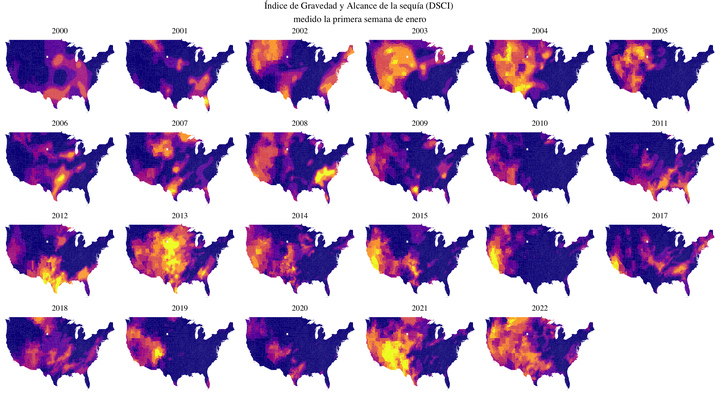
El tema de la semana 24 de #TidyTuesday es la sequía en EEUU y al leer sobre la cantidad de precipitación por año inmediatamente pensé que era una gran oportunidad para jugar con los raincloud plots. Sin embargo, los datos no estaban de mi lado, porque presentaban información temporoespacial. Así que decidí continuar con mi acercamiento al manejo de datos espaciales.
Por suerte unos meses atrás me había registrado en el curso de mapas coropléticos de Ari Lamstein, así que tenía material desde donde empezar.
Uno de los datasets del TidyTuesday tiene el Drought Severity and Coverage Index (DSCI) (algo así como el índice de gravedad y alcance de la sequía) junto con el código FIPS que indica el condado y el estado. Este código FIPS es el mismo que aparece con el nombre region en el mapa del paquete choroplethrMaps.
Entonces, cargo los paquetes necesarios y el mapa de los condados:
tuesdata <- tidytuesdayR::tt_load('2022-06-14')##
## Downloading file 1 of 2: `drought.csv`
## Downloading file 2 of 2: `drought-fips.csv`drought <- tuesdata$drought
drought_fips <- tuesdata$`drought-fips`
library(choroplethr)
library(choroplethrMaps)
library(tidyverse)
data("county.map")Ahora puedo renombrar en el dataframe de la sequía los valores correspondientes para poder usarlo como base para el mapa coroplético. También necesito especificar que el valor de DSCI (ahora llamado “value”) sea numérico, y no caracter. Con esto, ya puedo combinar el df de los datos geográficos de los condados (county.map) con mi df de los datos de sequía (drought.fips):
drought_fips <- drought_fips %>%
rename(region = FIPS,
value = DSCI)
drought_fips$region <- as.numeric(drought_fips$region)
county.map.drought <- county.map %>%
left_join(drought_fips, by = "region") %>%
select(long, lat, region, value, date, STATE, group)Solo resta generar uno por uno los gráficos para cada año: selecciono la fecha de la primera medición del año y elimino los estados de Alaska y las islas Hawaii (solo porque me dificulta el mapeo, no es nada personal):
# 2000
a2000 <- county.map.drought %>%
filter(date == "2000-01-04" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2000",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2001
a2001 <- county.map.drought %>%
filter(date == "2001-01-02" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2001",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2002
a2002 <- county.map.drought %>%
filter(date == "2002-01-01" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2002",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2003
a2003 <- county.map.drought %>%
filter(date == "2003-01-07" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2003",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2004
a2004 <- county.map.drought %>%
filter(date == "2004-01-06" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2004",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2005
a2005 <- county.map.drought %>%
filter(date == "2005-01-04" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2005",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2006
a2006 <- county.map.drought %>%
filter(date == "2006-01-03" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2006",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2007
a2007 <- county.map.drought %>%
filter(date == "2007-01-02" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2007",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2008
a2008 <- county.map.drought %>%
filter(date == "2008-01-01" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2008",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2009
a2009 <- county.map.drought %>%
filter(date == "2009-01-06" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2009",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2010
a2010 <- county.map.drought %>%
filter(date == "2010-01-05" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2010",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2011
a2011 <- county.map.drought %>%
filter(date == "2011-01-04" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2011",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2012
a2012 <- county.map.drought %>%
filter(date == "2012-01-03" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2012",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2013
a2013 <- county.map.drought %>%
filter(date == "2013-01-01" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2013",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2014
a2014 <- county.map.drought %>%
filter(date == "2014-01-07" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2014",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2015
a2015 <- county.map.drought %>%
filter(date == "2015-01-06" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2015",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2016
a2016 <- county.map.drought %>%
filter(date == "2016-01-05" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2016",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2017
a2017 <- county.map.drought %>%
filter(date == "2017-01-03" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2017",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2018
a2018 <- county.map.drought %>%
filter(date == "2018-01-02" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2018",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2019
a2019 <- county.map.drought %>%
filter(date == "2019-01-01" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2019",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2020
a2020 <- county.map.drought %>%
filter(date == "2020-01-07" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2020",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2021
a2021 <- county.map.drought %>%
filter(date == "2021-01-05" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2021",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))
# 2022
a2022 <- county.map.drought %>%
filter(date == "2022-01-04" & !STATE %in% c("02", "15")) %>%
ggplot(aes(long, lat, group=group)) +
geom_polygon(aes(fill = value))+
scale_fill_viridis_c(option = "plasma",
limits = c(0, 500))+
labs(title = "2022",
x = "", y = "")+
theme_void()+
theme(legend.position = "none",
plot.title = element_text(size = 12, hjust = 0.5,
family="Times"))Finalmente, los combino en un solo gráfico:
library(grid)
library(gridExtra)##
## Attaching package: 'gridExtra'## The following object is masked from 'package:dplyr':
##
## combine## The following object is masked from 'package:acs':
##
## combinegrid.arrange(grobs = list(a2000, a2001, a2002, a2003, a2004, a2005, a2006, a2007, a2008, a2009, a2010,
a2011, a2012, a2013, a2014, a2015, a2016, a2017, a2018, a2019, a2020,
a2021, a2022),
top = grid::textGrob("Índice de Gravedad y Alcance de la sequía (DSCI) \nmedido la primera semana de enero",
gp = gpar(fontsize = 14, fontfamily = "Times")),
nrow = 4)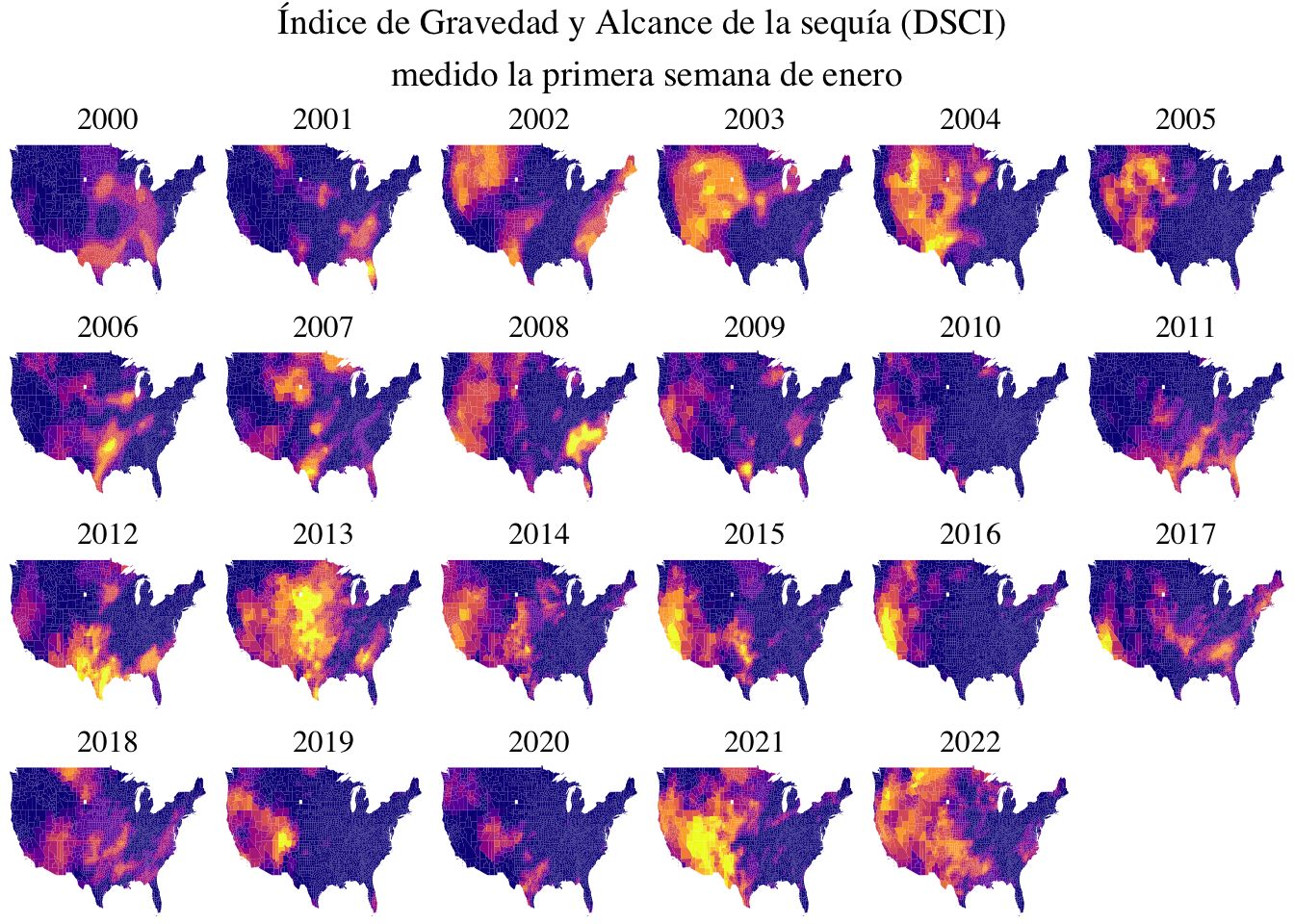
No estoy muy convencida de la relación de tamaños entre el título general y los títulos específicos de cada gráfico, pero por el momento estoy conforme con cómo se ve.
Ahora, todo muy lindo, pero ¿es informativo este gráfico? Decidí quedarme con la primera semana de enero por motivos arbitrarios, porque me interesaba enfocarme en lo espacial. Sin embargo, hubiera sido interesante por ejemplo hacer un promedio por mes a lo largo de los años (es decir, tener un mapa por mes que promediara los 22 años de la muestra), porque es evidente que la mayoría de las diferencias en los niveles de sequía se van a ver a lo largo del año (entre estaciones, digamos) que a lo largo de los años. Pero eso implicaba un manejo de los datos de fechas que todavía no tengo muy claro.